Abstract
Aromatic residues cluster in the core of folded proteins, where they stabilize the structure through multiple interactions. Nuclear magnetic resonance (NMR) studies in the 1970s showed that aromatic side chains can undergo ring flips—that is, 180° rotations—despite their role in maintaining the protein fold1,2,3. It was suggested that large-scale ‘breathing’ motions of the surrounding protein environment would be necessary to accommodate these ring flipping events1. However, the structural details of these motions have remained unclear. Here we uncover the structural rearrangements that accompany ring flipping of a buried tyrosine residue in an SH3 domain. Using NMR, we show that the tyrosine side chain flips to a low-populated, minor state and, through a proteome-wide sequence analysis, we design mutants that stabilize this state, which allows us to capture its high-resolution structure by X-ray crystallography. A void volume is generated around the tyrosine ring during the structural transition between the major and minor state, and this allows fast flipping to take place. Our results provide structural insights into the protein breathing motions that are associated with ring flipping. More generally, our study has implications for protein design and structure prediction by showing how the local protein environment influences amino acid side chain conformations and vice versa.
Main
Aromatic residues make up a considerable fraction of the hydrophobic core of folded proteins, where they stabilize the structure through CH–π (refs. 4,5,6), π–π (refs. 7,8) and cation–π (refs. 9,10) interactions as well as hydrogen bonds. NMR studies in the 1970s demonstrated that aromatic side chains can undergo ring flips—that is, 180° rotations of the χ2 dihedral angle (Cβ–Cγ axis)—even when engaged in stabilizing interactions in the hydrophobic core1,2,3. These ring flips require concerted movements of the surrounding residues (large-amplitude protein breathing motions), and ring flipping rates as a function of temperature and pressure have been used to report on these motions by deriving activation energies and volumes11,12,13,14,15,16,17,18,19,20,21. However, the structural details of ring flipping and the associated breathing motions have remained unknown, probably owing to difficulties in stabilizing ring flipping transition states or intermediates that are amenable to structure elucidation.
Here we capture ring flipping events of a buried tyrosine residue in the SH3 domain of the JNK-interacting protein 1 (JIP1). We show using NMR relaxation dispersion that the aromatic ring of this tyrosine residue populates a minor-state conformation (3%), and we design single point mutations to stabilize this conformation and capture its high-resolution structure using X-ray crystallography. The structure reveals how the intricate network of hydrogen bonds and CH–π interactions is rearranged in the minor state. We show how a substantial void volume is generated around the tyrosine ring during the structural transition from the major to the minor state, which can be associated with the breathing motions that allow fast-timescale ring flipping events to take place. Our results provide structural insights into aromatic ring flipping and its associated protein breathing motions.
Protein dynamics induced by a tyrosine residue
The SH3 domain of JIP1 undergoes exchange between two distinct conformations, as evidenced by 15N NMR relaxation measured at multiple temperatures (Extended Data Figs. 1, 2, Supplementary Discussion). To analyse the observed dynamics in detail, we acquired 15N and 1HN Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion experiments at 15 °C (refs. 22,23,24,25). These experiments quantify the kinetics of exchange processes and provide the difference in chemical shift between a major and a minor state, together with their relative populations26,27,28,29,30. The data confirm that exchange contributions to the transverse relaxation are present for residues within three regions of the protein (Extended Data Fig. 1d, e, Supplementary Table 1). These residues are located spatially close to tyrosine 526 (Y526) (Extended Data Fig. 1f). A mutation of Y526 to alanine (Y526A) shows no conformational exchange (Extended Data Fig. 1d, e), and conserves the protein backbone conformation, as evidenced from its crystal structure that we obtained at 1.5-Å resolution (Extended Data Fig. 3, Extended Data Tables 1, 2). These results show that the relaxation dispersion that affects around 40% of the residues in the SH3 domain arises from a single exchange process, with Y526 being the origin of the observed exchange.
The 15N and 1HN relaxation dispersion data were analysed simultaneously according to a two-site exchange model in which a highly populated major state interconverts with a low-populated minor state (Extended Data Fig. 4a, b, Supplementary Discussion). The analysis of the data gives a population of the minor state of pminor = 2.8 ± 0.1% and an exchange rate constant of kEX = 2,600 ± 70 s−1. The derived chemical shift differences, ΔδCPMG, span a range of 4.7 ppm for 15N and 1.1 ppm for 1HN suggesting that there are substantial structural changes between the major and the minor state (Extended Data Fig. 4c, d).
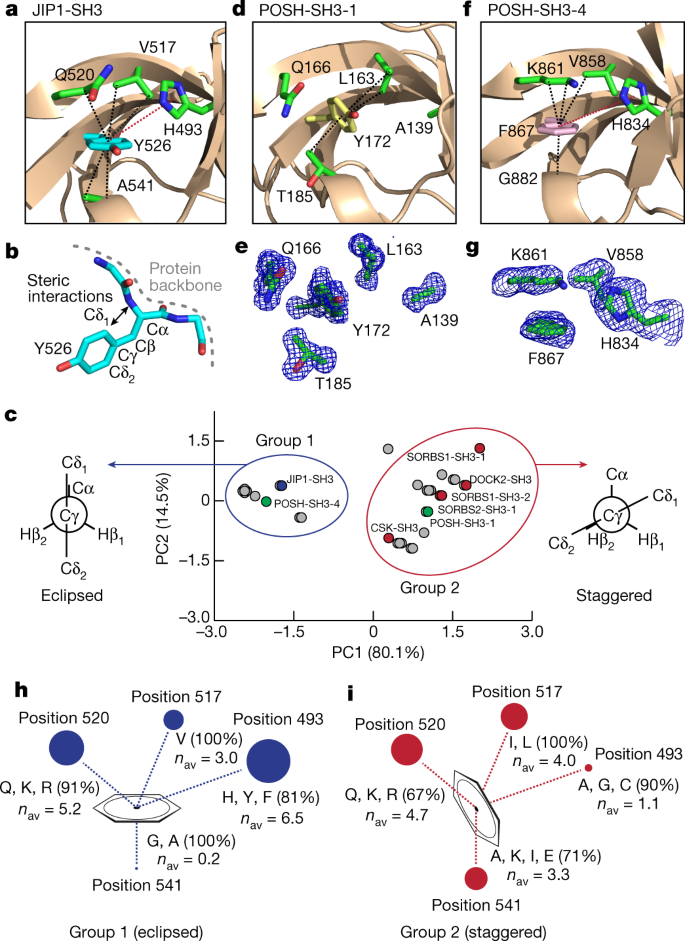
The side chain of Y526 is found in an unusual conformation in the crystal structure (Protein Data Bank (PDB) code 2FPE (ref. 31), Extended Data Table 1), characterized by a χ2 dihedral angle of 2° (Fig. 1a). Normally, this eclipsed conformation is energetically unfavourable because of steric interactions with the backbone (Fig. 1b), and it is rarely found in proteins as χ2 angles are preferred where Cδ1 and Cδ2 are staggered with respect to Cα (ref. 32). However, the eclipsed conformation of the aromatic ring of Y526 is stabilized by CH–π interactions from V517, Q520 and A541 and by π–π interactions with H493 (Fig. 1a).
a, Crystal structure of wild-type JIP1-SH3, showing the conformation of Y526 and its stabilizing interactions with H493, V517, Q520 and A541. Dashed lines indicate CH–π (black) and π–π (red) interactions. b, Side chain conformation of Y526 in JIP1-SH3, illustrating the steric interactions between the δ1 nuclei of the aromatic ring and the backbone. c, PCA of a dataset comprising the size of the amino acid side chains at positions 493, 517 and 541 within SH3 domains that contain Y or F at position 526 (Extended Data Fig. 5b). Two groups are observed, which correspond to eclipsed (group 1) or staggered (group 2) conformations of the aromatic ring. JIP1-SH3 is indicated in blue; POSH-SH3-1 and POSH-SH3-4 are indicated in green; and SH3 domains for which crystal structures have been determined previously are shown in red. d, Crystal structure of POSH-SH3-1, showing a staggered conformation of Y172. e, Unbiased electron density maps (Fo–Fc) of Y172 and the surrounding residues in POSH-SH3-1. f, Crystal structure of POSH-SH3-4, showing an eclipsed conformation of F867. g, Unbiased electron density maps (Fo–Fc) of F867 and the surrounding residues in POSH-SH3-4. h, i, Results of the PCA, illustrating the size and nature of the residues that surround the aromatic ring in position 526 in group 1 (h) and group 2 (i) SH3 domains. The size of the spheres in each position is proportional to the average size (nav) of the amino acid side chain across group members.
Proteome-wide SH3 sequence analysis
To investigate the contribution from the surrounding residues in stabilizing the eclipsed conformation of Y526, we analysed the sequences of all identified human SH3 domains33. We categorized the sequences according to the identity of the amino acid at the position of Y526 in the JIP1 SH3 domain (JIP1-SH3) (Extended Data Fig. 5a), and we retained the sequences carrying a phenyl-based amino acid (Tyr or Phe) at this position, amounting to 33 SH3 domains. Sequence alignments reveal a large variation in the size of the amino acids at positions 493, 517 and 541, whereas at position 520 most sequences contain Gln, Arg or Lys (72%) (Extended Data Fig. 5b, c). To study the size correlation between the amino acids at positions 493, 517 and 541 and their influence on the conformation of the aromatic residue at position 526, we carried out a principal component analysis (PCA) by assigning a size score (n) to each amino acid according to the number of heavy atoms in their side chains. This analysis reveals two well-separated groups, with the SH3 domain of JIP1 belonging to group 1 (Fig. 1c). In group 2, five SH3 domains are found for which high-resolution crystal structures are available; these include three SH3 domains of the sorbin and SH3 domain-containing proteins 1 and 2 (SORBS1 and SORBS2)34, and the SH3 domains of the dedicator of cytokinesis protein 2 (DOCK2)35 and of the tyrosine protein kinase CSK36 (Extended Data Fig. 5b). Notably, all group 2 structures show a favourable, staggered side chain conformation (of Cδ1/Cδ2 with respect to Cα) of the corresponding tyrosine, with the χ2 dihedral angle ranging from −40° to −64° (Fig. 1c). We therefore hypothesized that SH3 domains of group 1 have eclipsed conformations, whereas group 2 have staggered conformations. To test our hypothesis, we determined two crystal structures of SH3 domains of the scaffold protein POSH (‘plenty of SH3 domains’), for which the first SH3 domain belongs to group 2 and the fourth SH3 domain belongs to group 1 (Extended Data Table 2, Fig. 1c). Consistent with the PCA, the crystal structure of the first SH3 domain of POSH (POSH-SH3-1) shows a staggered conformation of the corresponding tyrosine residue (Y172), which is stabilized by CH–π interactions from L163 (position 517) and T185 (position 541) (Fig. 1d, e). POSH-SH3-4 shows a similar structure to JIP1-SH3, with an eclipsed conformation of the corresponding phenylalanine residue (F867) stabilized by CH–π interactions from V858 (position 517), K861 (position 520) and G882 (position 541), and by π–π interactions with H834 (position 493) (Fig. 1f, g). Our data therefore suggest that the conformation of the aromatic ring at position 526 is determined by steric interactions dictated by the size of the surrounding amino acids.
Structure of the minor state
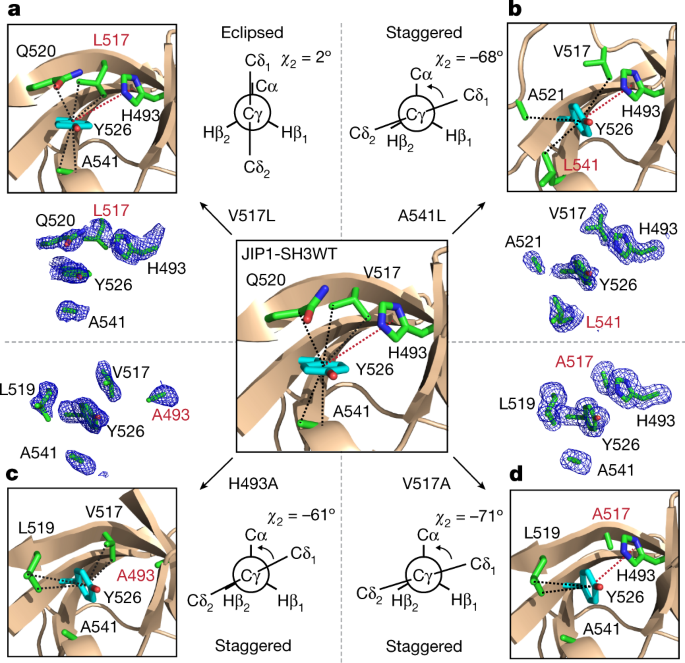
Next, we investigated whether the minor state detected by NMR corresponds to a staggered conformation of the side chain of Y526. We sought to stabilize the minor state relative to the major state by introducing single point mutations. The PCA suggests that a staggered conformation in group 2 is favoured over an eclipsed conformationin group 1 when residues with larger side chains are found in position 541, when isoleucine or leucine are occupying position 517 and when smaller residues are found in position 493 (Fig. 1h, i). Accordingly, we designed four different mutants of JIP1-SH3 (H493A, V517A, V517L and A541L) and obtained their high-resolution structures by X-ray crystallography (Extended Data Tables 1, 2). Of note, three mutants (H493A, V517A and A541L) induce a staggered conformation of Y526, with χ2 dihedral angles ranging from −41° to −75°, whereas V517L shows an eclipsed conformation of Y526 and an almost identical structure to the wild-type protein (Fig. 2, Extended Data Fig. 6a). The high resolution of these structures, ranging from 1.4 to 1.9 Å, allows unambiguous determination of the conformation of Y526 and the surrounding residues, as demonstrated by their unbiased electron density maps (Fig. 2).
a–d, Crystal structures showing the conformation of Y526, the corresponding unbiased electron density maps (Fo–Fc) of Y526 and its surrounding residues, and the Newman projection along the Cβ–Cγ bond of Y526 in the V517L (a), A541L (b), H493A (c) and V517A (d) variants of JIP1-SH3. Dashed lines indicate CH–π (black) and π–π (red) interactions. Residue numbers in red indicate the site of mutation. The wild-type JIP1-SH3 structure is shown as a reference in the centre.
Structural details of breathing motions
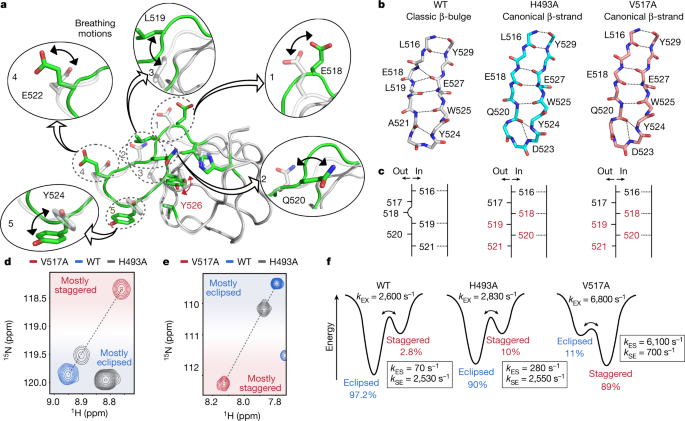
The two variants H493A and V517A show almost identical crystal structures (Extended Data Fig. 6b) and an equivalent stabilization mechanism of the aromatic ring of Y526. Whereas the wild-type structure exhibits a classic β-bulge at residue 518 (ref. 37), the transition from the eclipsed to the staggered conformation induces a local inversion (in-out) at residues 518–520, which leads to the formation of a canonical β-strand, as observed in the structures of the H493A and V517A variants (Fig. 3a–c). This transition allows the side chain of L519 to rearrange and form CH–π interactions with the ring of Y526 (Fig. 2c, d); and, at the same time, large-scale movements of E518, Q520, E522 and Y524 are observed (Fig. 3a). We note that SH3 domains in both group 1 and group 2 of the PCA show classic β-bulges at position 518 (Extended Data Fig. 5d), which suggests that the presence of this structural motif is not determinant of the side chain conformation of the phenyl ring in position 526.
a, Structural changes associated with the transition from the eclipsed (green) to the staggered (grey) conformation of Y526. b, Illustration of the backbone conformation of the β-sheet formed between the 516–521 and 524–529 regions in the wild-type (WT) protein (left), and in the H493A (middle) and V517A (right) variants. Dashed lines indicate hydrogen bonds. c, Schematic representation of the conformation of the β-strand encompassing residues 516–521, showing the orientation of the carbonyl group (‘out’, carbonyl group surface exposed; ‘in’, carbonyl group pointing towards the β-strand encompassing residues 524–529) in the wild-type protein (left) and in the H493A (middle) and V517A (right) variants. d, e, Two examples of linear correlations between the chemical shifts of wild-type JIP1-SH3 (blue spectrum) and the two variants H493A (grey spectrum) and V517A (red spectrum) as observed in 1H–15N HSQC spectra acquired at 35 °C (d, residue E522; e, residue D524). f, Energy landscapes illustrating the effect of single point mutations on the exchange rate constants and fractional populations of the major (eclipsed) and minor (staggered) conformations as determined by relaxation dispersion experiments acquired at 15 °C.
The A541L mutation also triggers a staggered conformation of Y526 and a rearrangement of the 517–522 region; however, the stabilization mechanism of the aromatic ring is different. A looping out of the β-strand between residues 517 and 522 is observed (Extended Data Fig. 6c–e), which allows the side chain of A521 to reorient and to stabilize the staggered conformation of the ring through CH–π interactions together with L541 and V517 (Fig. 2b). At the same time, the side chains of L519, Q520 and E522 and D523 undergo large-scale movements to accommodate the flipped ring within the pocket (Extended Data Fig. 6c).
Altogether, the different mutants show that the dynamics of the region encompassing residues 517–522 are key for the formation of the minor state. The experimental 13C chemical shifts for residues in this region are characteristic of random coil conformations (Extended Data Fig. 1b) and, compared to other regions of secondary structure, the relaxation-derived order parameters (S2) are lower (Extended Data Fig. 2g) and the crystallographic B-factors are higher. This supports the idea of the 517–522 region being intrinsically dynamic, with a fluctuating hydrogen-bonding network that is prone to structural transitions.
Next, we sought to determine which of the two crystal structures (H493A/V517A-like or A541L-like) best capture the conformation of the wild-type minor state detected by NMR relaxation dispersion. The H493A and V517A variants show almost identical crystal structures and for a subset of residues, the chemical shifts of which are unaffected by the mutations, the resonances of the two variants fall on a straight line together with the resonances of the wild-type protein (Fig. 3d, e). This suggests that they are in fast–intermediate exchange between two conformations represented by the crystal structures of the wild-type protein and of the H493A/V517A variants. In agreement with this, both variants show line broadening and chemical exchange contributions as detected by 15N and 1HN relaxation dispersion (Extended Data Figs. 7a–c, 8a–c). Analysis of these data (Extended Data Figs. 7d, e, 8d, e) shows that the structural features of the minor state of the wild-type protein are captured by the H493A/V517A crystal structures, as shown by the excellent agreement between the ΔδCPMG values for the wild-type protein and the two variants (Extended Data Figs. 7f, g, 8f, g). In addition, the analysis yields exchange rates between the staggered (canonical β-strand) and the eclipsed (classic β-bulge) conformation of kEX = 2,830 ± 70 s−1 (H493A) and kEX = 6,800 ± 300 s−1 (V517A), compared to kEX = 2,600 ± 70 s−1 determined for the wild-type protein. Finally, the observable chemical shifts of the two variants (Fig. 3d, e), in conjunction with analysis of the relaxation dispersion data (Supplementary Discussion), show that the H493A mutation slightly stabilizes the minor state relatively to the major state, whereas the V517A mutation almost inverts the relative populations of the major and minor states (Fig. 3f). For completeness, we note that the A541L crystal structure is not representative of the minor state conformation (Extended Data Fig. 9), although it shares structural features that are necessary for fast ring flipping of Y526 (see below).
Void volume enables ring flipping
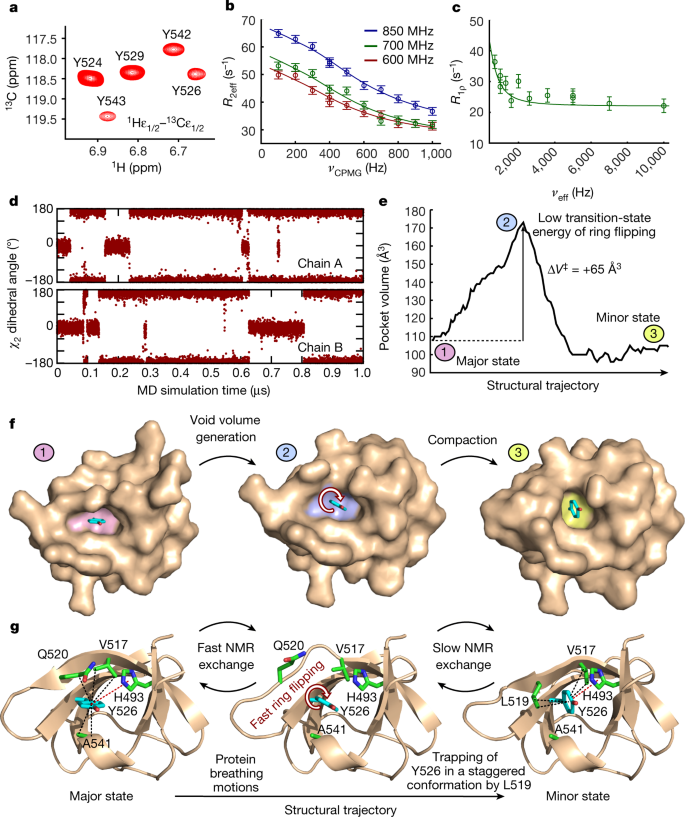
Aromatic 1H–13C heteronuclear single quantum coherence (HSQC) spectra show averaging of the NMR signals of the tyrosine ε1/ε2 nuclei of Y526 (Fig. 4a, Extended Data Fig. 10a, Supplementary Fig. 1), which shows that full 180° ring flipping occurs. This poses the question of the timescale of the full ring flipping and its relation to the observed minor state. To answer this question, we acquired L-optimized TROSY-selected aromatic side chain 13Cε CPMG (ref. 38) and on-resonance R1ρ (ref. 39) relaxation dispersion data of Y526. These data are entirely explained by the exchange process between the major and minor state, with a negligible contribution from the full ring flipping event (Fig. 4b, c), demonstrating that ring flipping of Y526 is fast (kEX > 50,000 s−1) (Supplementary Discussion). This observation agrees with a 1-μs molecular dynamics (MD) simulation that shows several 180° ring flipping events of Y526 (Fig. 4d, Extended Data Fig. 10b, c).
a, 1H–13C HSQC spectrum of JIP1-SH3 at 15 °C showing tyrosine epsilon correlations. b,13Cε CPMG relaxation dispersion profiles of Y526 obtained at 15 °C (red, 600 MHz; green, 700 MHz; blue, 850 MHz). The data were analysed simultaneously according to a two-site exchange model (full-drawn lines). Error bars represent one s.d. derived from Monte Carlo simulations of experimental uncertainty. c, 13Cε on-resonance R1ρ relaxation dispersion profile of Y526 at 15 °C and 700 MHz. The full-drawn line corresponds to the calculated R1ρ profile from the exchange parameters (kEX, pminor and ΔδCPMG) obtained from the analysis of the CPMG data in b. Error bars represent one s.d. and were derived as in b. d, The dihedral angle χ2 of Y526 as a function of simulation time in a 1-μs MD simulation of the dimeric JIP1-SH3. e, Volume of the pocket of Y526 across the structural trajectory between the major and minor state. f, Surface representation of JIP1-SH3 in three different states corresponding to the major state, an intermediate state on the structural trajectory and the minor state. The Y526 pocket is highlighted in pink (major), blue (intermediate) and yellow (minor). The rearrangements along the structural trajectory between the major and the minor state generate a void volume around Y526, thereby lowering the transition-state energy of ring flipping. g, Illustration of the protein breathing motions along the structural trajectory from the major to the minor state. A void volume is created around Y526, which allows fast ring flipping to take place. The ring flipping is occasionally interrupted by trapping of Y526 in a staggered conformation through formation of CH–π interactions with L519 enabled by the β-bulge to β-sheet transition.
During the structural transition between the major and the minor state (Supplementary Video 1), a void volume is created around the ring of Y526 that corresponds to a pocket expansion of 65 Å3; this is mainly due to the structural reorganization of the side chain of Q520 (Fig. 4e, f). This cavity expansion is in agreement with previous studies that have reported activation volumes between 40 and 85 Å3 for ring flipping events of aromatic residues in other proteins12,14,21,40,41. The expansion is followed by a compaction of the surrounding protein environment as the ring becomes stabilized by CH–π interactions from L519 (Fig. 4e, f).
Collectively, our results are consistent with a model in which fast protein breathing motions along the structural trajectory between the major and the minor state generate the necessary void volume for ring flipping to take place by lowering the energy of the transition state (Fig. 4e–g, Supplementary Video 2). Occasionally, the β-bulge to β-sheet transition is completed and the aromatic ring becomes trapped in a staggered conformation that is stabilized by CH–π interactions with L519—a process that gives rise to the observed relaxation dispersion. These events are rare and occur on a slow timescale (Figs. 3f, 4g), but they constitute an important tool for observing the trajectory of the protein breathing motions coupled to aromatic ring flipping. The initial generation of void volume around the ring is almost identical along the structural trajectory between the major state (wild type) and the A541L crystal structure (Extended Data Fig. 6f). Thus, all mutants—including A541L, which stabilizes Y526 in a staggered conformation by a different mechanism—share the same initial structural trajectory and report on identical breathing motions.
Conclusions
Our results provide structural insights into the protein breathing motions that are associated with aromatic ring flipping. We reveal how the dynamics of the region encompassing residues 517 to 522 are key for accommodating the ring flipping process of Y526. Notably, the transition from the eclipsed, major conformation to the staggered, minor conformation is associated with a structural change from a rare, classic β-bulge to a common, canonical β-strand conformation (Supplementary Video 1, Fig. 3a–c). Breathing motions along the structural trajectory between the major and the minor state generate the necessary void volume for fast ring flipping of Y526 to take place (Supplementary Video 2). Although a recent NMR study suggested extensive local unfolding as the source of cavity creation41, our study provides an alternative view by showing how a substantial void volume can be generated through distinct structural rearrangements, while maintaining the overall protein fold.
More generally, our study shows how the local environment in the protein core can lead to a priori energetically unfavourable conformations of amino acid side chains, and how subtle changes in this environment can lead to major structural rearrangements to revert to preferred amino acid conformations. Our results therefore have implications for protein design and structure prediction, and for how novel biological functions can be acquired during the course of evolution; for example, by altering the delicate balance between hydrogen bonds and CH–π interactions. Finally, the combination of sensitive NMR methods to detect low-populated states, protein design using proteome-wide sequence analyses and high-resolution crystallography could be a strategy to further discover the structural details of sparsely populated protein states and their link to function.
Methods
Expression and purification of JIP1-SH3, POSH-SH3-1 and POSH-SH3-4
JIP1-SH3, corresponding to residues 490–549 of human JIP1 (Uniprot Q9UQF2), was subcloned into a pET28a vector, and two of the four SH3 domains of the E3 ubiquitin-protein ligase SH3RF1 (Uniprot Q7Z6J0; also known as ‘plenty of SH3 domains’ (POSH)), SH3-1 (135–194) and SH3-4 (829–888), were subcloned into a pESPRIT vector42. The constructs therefore contained an N-terminal hexahistidine tag followed by a tobacco etch virus (TEV) cleavage site. The final proteins after protease cleavage contain N-terminal GRR (POSH) or GHM extensions (JIP1).
To obtain unlabelled proteins, Escherichia coli BL21(DE3) cells transformed with one of the constructs were grown in lysogeny broth (LB) medium at 37 °C until the optical density at 600 nm reached 0.7. Protein expression was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cultures were grown for an additional 15 h at 20 °C (POSH-SH3s) or 4 h at 37 °C (JIP1-SH3). The cells were collected by centrifugation and frozen at −20 °C or −80 °C. Isotopically 15N/13C- and 15N-labelled proteins were produced by growing transformed E. coli BL21(DE3) cells in M9 minimal medium containing 1 g l−1 of 15N-NH4Cl and 2 g l−1 of 13C6-d-glucose or 12C6- d-glucose. To obtain 15N-labelled protein with tyrosine residues site-selectively labelled at the ε position with 13C, transformed E. coli BL21(DE3) cells were grown in M9 minimal medium containing 1 g l−1 of 15N-NH4Cl, 2 g l−1 of NaH13CO3 and 2 g l−1 of [2-13C]-glycerol43.
All SH3 domains were purified by Ni affinity chromatography followed by size-exclusion chromatography. Cell lysis was carried out by sonication using purification buffer (POSH: 50 mM Tris pH 7.0/8.0, 500 mM NaCl, 1 mM β-mercaptoethanol; JIP1: 50 mM HEPES pH 7.0, 150 mM NaCl) supplemented with protease inhibitor tablets (Roche). The washing buffer used for Ni affinity chromatography was the same as the purification buffer with the addition of 20 mM imidazole. The elution buffer was the same as the purification buffer with the addition of 500 mM (POSH) or 300 mM (JIP1) imidazole. Nickel affinity chromatography was followed by cleavage by the TEV protease, a second Ni affinity column and size-exclusion chromatography on a Superdex 75 (GE Healthcare). This column was equilibrated with 50 mM HEPES pH 8.0, 500 mM NaCl, 2 mM DTT for POSH-SH3-1, 50 mM HEPES pH 7.0, 500 mM NaCl, 2 mM DTT for POSH-SH3-4 and 50 mM HEPES pH 7.0, 150 mM NaCl for JIP1-SH3.
Expression and purification of JIP1-SH3 variants
Expression and purification of the JIP1-SH3 variants (Y526A, V517A, V517L, A541L and H493A) were performed following the same protocol as for JIP1-SH3, except that the cultures were grown for 15 h after induction at 20 °C (instead of 4 h at 37 °C).
Thermal stability measurements of JIP1-SH3
The stability of JIP1-SH3 was measured by differential scanning fluorimetry using a Prometheus NT.48 (Nanotemper) instrument with the emission wavelengths set to 330 and 350 nm and an excitation power of 10%. The melting curve for wild-type JIP1-SH3 was measured at a protein concentration of 4 mg ml−1 in 50 mM HEPES, 150 mM NaCl at pH 7.0 by using Prometheus Standard Capillaries (PR-C002). The temperature scan rate was fixed at 1 °C per min from 20 °C to 95 °C. The melting temperature (Tm) was calculated from the peak of the first derivative of the intrinsic protein fluorescence intensity ratio at 350 nm and 330 nm throughout the duration of the temperature ramp.
NMR spectral assignment of JIP1-SH3 and its variants
The NMR assignment experiments were acquired in 50 mM HEPES, 150 mM NaCl, pH 7.0 at a protein concentration of 0.94 mM (JIP1-SH3), 1.06 mM (JIP1-SH3(Y526A)), 1.10 mM (JIP1-SH3(A541L)), 2 mM (JIP1-SH3(V517A)) and 0.90 mM (JIP1-SH3(H493A)). The NMR spectral assignments of JIP1-SH3 were performed at 25 °C using a set of BEST-TROSY triple resonance experiments (HNCO, intra-residue HNCACO, HNCOCA, intra-residue HNCA, HNCOCACB and intra-residue HNCACB) acquired at a 1H frequency of 600 MHz (Bruker, operated with TopSpin v.3.5)44. The NMR spectral assignments of JIP1-SH3(Y526A) were obtained at 25 °C at a 1H frequency of 700 MHz (Bruker) using BEST-TROSY HNCO, HNCOCACB and intra-residue HNCACB experiments. The NMR spectral assignments of JIP1-SH3(A541L) (at 25 °C), JIP1-SH3(V517A) (at 35 °C) and JIP1-SH3(H493A) (at 35 °C) were obtained at a 1H frequency of 700 MHz (Bruker) using a BEST-TROSY HNCACB experiment. The spectra were manually peak-picked using NMRFAM-Sparky45 and sequential connectivities were identified manually or by using the assignment program MARS46. Secondary structure propensities were calculated using SSP on the basis of the experimental Cα and Cβ chemical shifts47.
15N relaxation measurements of JIP1-SH3
Measurements of 15N relaxation rates (R1, R2 and heteronuclear NOEs) of JIP1-SH3 were obtained using standard HSQC-type pulse sequences48 at a 1H frequency of 600 MHz (Agilent, operated with VnmrJ v.3.1). The relaxation rates were measured at four different temperatures: 15, 25, 35 and 45 °C. The magnetization decay was sampled at (0, 100, 200, 400, 600, 800, 1,100, 1,500 and 1,900) milliseconds (ms) for longitudinal and at (10, 30, 50, 70, 90, 130, 170, 210 and 250) ms for transverse relaxation. Technical replicates of one or two of these delays were acquired to estimate the uncertainty on the relaxation rates using a Monte Carlo approach. Details of the Lipari–Szabo model free analysis can be found in the Supplementary Discussion.
15N and 1HN CPMG relaxation dispersion of JIP1-SH3 and its variants
All 15N CPMG relaxation dispersion experiments24 were carried out at 15 °C using a constant-time relaxation delay of 32 ms with CPMG frequencies (νCPMG) ranging from 31.25 to 1,000 Hz and a 1H decoupling field of 11 kHz. The 1HN relaxation dispersion experiments were carried out at 15 °C using the published pulse sequence25 with a constant-time relaxation delay of 20 ms and CPMG frequencies ranging from 50 to 2,000 Hz. Uncertainties on peak intensities extracted from the relaxation dispersion experiments were estimated using the pooled s.d. calculated from repeat measurements (technical replicates of one to three νCPMG values), each pool being the set of repeat points per νCPMG and per peak. Uncertainties on R2eff values were propagated from the peak intensity uncertainty using a Monte Carlo approach. The following relaxation dispersion experiments were acquired: JIP1-SH3: 15N (600 MHz, Agilent), 15N (850 MHz, Bruker), 1HN (600 MHz, Bruker), 1HN (950 MHz, Bruker); JIP1-SH3(H493A) and JIP1-SH3(V517A): 15N (600 MHz, Bruker), 15N (950 MHz, Bruker), 1HN (600 MHz, Bruker), 1HN (950 MHz, Bruker); JIP1-SH3(Y526A): 15N (700 MHz, Bruker) and 1HN (600 MHz, Bruker); JIP1-SH3(A541L): 15N (700 MHz, Bruker). All relaxation dispersion data were analysed using the program ChemEx (https://github.com/gbouvignies/ChemEx)49 as described in the Supplementary Discussion.
Tyrosine assignments and 13C CPMG relaxation dispersion of Y526
The 13Cε–1Hε tyrosine resonances were assigned at 45 °C by acquiring a two-dimensional (2D) plane of a BEST-TROSY intra-residue HNCACB experiment44, an aromatic BEST constant-time 1H–13C HSQC experiment50 and a (Hβ)Cβ(CγCδCε)Hε experiment51 linking the Cβ chemical shifts directly to the Hε chemical shifts. The spectra were manually peak-picked using NMRFAM-Sparky45 and 13Cε–1Hε tyrosine resonances were assigned manually (Supplementary Fig. 1). The acquisition of aromatic BEST constant-time 1H–13C HSQC experiments at different temperatures (between 5 and 45 °C) enabled the final assignment at 15 °C.
Aromatic L-optimized TROSY-selected 13C CPMG38 and R1ρ39 relaxation dispersion experiments were carried out at 15 °C on a 1 mM uniformly 15N and site-selective 13C-labelled JIP1-SH3 sample in 50 mM HEPES, 150 mM NaCl at pH 7.0. CPMG relaxation dispersion experiments were carried out at magnetic field strengths of 600 MHz, 700 MHz and 850 MHz (Bruker) using a constant-time relaxation delay of 20 ms with CPMG frequencies ranging from 100 to 1,000 Hz. The R1ρ relaxation dispersion experiment was recorded on-resonance with Y526 at 700 MHz (Bruker) using B1 field strengths ranging from 700 to 10,000 Hz with a 20 ms relaxation delay. Error bars were derived from repeat measurements as described above for the 15N and 1HN relaxation dispersion experiments. Analysis of the 13C data was carried out using ChemEx or using available analytical expressions for R1ρ relaxation in the presence of two-site exchange52 (Supplementary Discussion).
Comparison of JIP1-SH3 to other human SH3 domains
The sequences of 320 human SH3 domains were obtained33, aligned using Clustal Omega53 and categorized according to the identity of the amino acid at the position of Y526 in JIP1-SH3. A new alignment was performed using only the sequences that carry Y or F at this position, amounting to a total of 33 human SH3 domains. The sequences of the SH3 domains of the RIMS-binding proteins 1, 2 and 3 and the metastasis-associated in colon cancer protein 1 (MACC1) were not included in this alignment, as they contain longer insertions compared to the JIP1-SH3 sequence. For each SH3 domain, the amino acids corresponding to residues 493, 517 and 541 of JIP1-SH3 were assigned a size score according to the number of heavy atoms in their side chains (A: 1, C: 2, D: 4, E: 5, F: 7, G: 0, H: 6, I: 4, K: 5, L: 4, M: 4, N: 4, P: 3, Q: 5, R: 7, S: 2, T: 3, V: 3, Y: 8, W:10). A PCA was carried out to reveal potentialcorrelations between the sizes of the amino acids in position 493, 517 and 541 using the ClustVis webtool54.
Crystallization of JIP1-SH3 and the variants Y526A, V517A, V517L, A541L and H493A
JIP1-SH3 and its variants were concentrated to a final concentration of 20 mg ml−1 (JIP1-SH3, JIP1-SH3(V517A), JIP1-SH3(A541L) and JIP1-SH3(V517L)), 10 mg ml−1 (JIP1-SH3(Y526A)) and 4 mg ml−1 (JIP1-SH3(H493A)) after size-exclusion chromatography by using Amicon Ultra-4 3.0-kDa centrifugal filters (Merck). All crystals were obtained in 0.1 M HEPES pH 7.5, 1–5% PEG 400 and 2–2.5 M ammonium sulfate at 20 °C by the hanging-drop vapour diffusion method in 24-well plates (Hampton research)55. Drops of 2–3 μl consisting of 1:1 or 2:1 parts of protein solution and reservoir solution were vapour-equilibrated against 500 μl of reservoir solution. All crystals appeared after two days and were collected by transferring them to a mother liquor solution containing 20–30% trehalose, frozen and kept in liquid nitrogen.
Crystallization of POSH-SH3-1 and POSH-SH3-4
Purified POSH-SH3-1 and POSH-SH3-4 were directly concentrated after size-exclusion chromatography to 3.4 and 5.0 mg ml−1, respectively, using Amicon Ultra-4 3.5-kDa centrifugal filters (Merck). Initial crystallization conditions were identified using the high-throughput crystallization platform (EMBL).
The initial condition identified for POSH-SH3-1 was 0.2 M NaF, 20% PEG 3350 from the PEGs-I screen (Qiagen) at 4 °C. Needles appeared after 3 to 7 days. Further optimization was done using the hanging-drop vapour diffusion method at 4 °C in 24-well plates. Drops of 2 μl consisting of equal parts protein solution at 2.5 mg ml−1 and reservoir solution (0.2 M NaF, 22% PEG 3350) were vapour-equilibrated against 500 μl of reservoir solution. Hexagonal crystals appeared after three days and were collected after five days by transferring them to a mother liquor solution containing 5% ethylene glycol as cryoprotectant, frozen and kept in liquid nitrogen.
The initial screen of POSH-SH3-4 identified two crystallization conditions: 0.1 M MES pH 6.5, 25% PEG 3000 (condition 1) and 0.1 M MES pH 6.5, 25% PEG 4000 (condition 2) from the PEGs-I screen (Qiagen) at 4 °C. Diffraction-quality needles (condition 1, 0.1 M MES pH 6.5, 26% PEG 3000) or three-dimensional crystals (condition 2, 0.1 M MES pH 6.5, 23% PEG 4000) were obtained after four days using the same vapour-diffusion set-up as for POSH-SH3-1. These were collected after seven days with 10% ethylene glycol as cryoprotectant, frozen and kept in liquid nitrogen.
Structure determination
Crystal diffraction was performed at the ESRF beamlines ID30A, ID23-1, ID23-2 using the MXCube software56,57, at the automated beamline MASSIF-158 or at the Diamond beamlines I04 and I04-1, all equipped with Pilatus detectors (Dectris). Indexing and integration was performed using the XDS59, the autoProc60 or GrenADeS61 program suites. Data reduction for JIP1-SH3(H493A) was carried out with Pointless and Aimless62,63. Molecular replacement of the wild-type JIP1-SH3 structure was carried out in Phaser64 using the PDB code 2FPE (chains A–B) as a search model. The structures of JIP1-SH3 mutants were obtained by using our wild-type JIP1-SH3 structure as a search model. The initial solutions were improved through cycles of manual adjusting in Coot65 and refined by using Refmac566. Aimless, Phaser and Refmac were all used as programs of the CCP4 suite67.
The structure of POSH-SH3-4 was determined by molecular replacement using a homology model that was built on the basis of the SH3 domain structure of SORBS1 (PDB code: 2LJ1, chain A), which has 45% sequence identity. The structure of POSH-SH3-1 was determined by molecular replacement using as a search model the SH3 domain of human tyrosine protein kinase C-Src (PDB code: 2SRC). Crystallography applications were compiled and configured by SBGrid68.
Structural trajectory and void volume calculations
The structural trajectory between the major and minor conformation was generated with Chimera69 by morphing between the wild-type JIP1-SH3 structure (PDB 7NYK) and the structures of the two variants JIP1-SH3(H493A) (7NYL) and JIP1-SH3(V517A) (7NYM). To calculate changes in the volume of the Y526 pocket, protons were added to all structures of the trajectory and Y526 was replaced by glycine to allow calculation of the complete pocket volume by POVME 3.0 using a distance cut-off of 1.09 Å, corresponding to the van der Waals radius of a hydrogen atom70. A similar strategy was used to generate the structural trajectory between the wild-type JIP1-SH3 structure (PDB 7NYK) and the structure of the JIP1-SH3(A541L) variant (7NYO).
MD simulations of JIP1-SH3
MD simulations were carried out using ACEMD v.3.3.071 and the Charmm36m force field parameters72. Using VMD73, coordinates of the dimer from PDB 2FPE were inserted in the box of dimensions with a minimum distance of 2 Å in each direction between each atom and any box side. The box was then filled with water molecules and an amount of Na+ and Cl− corresponding to [NaCl] = 0.1 M. Electrostatic interactions were evaluated using Particle-Mesh Ewald (PME) electrostatics with a cut-off distance of 9 Å. Van der Waals forces were calculated with a cut-off of 9 Å and a switching function active from 7.5 Å to smoothly reduce the potential to zero. An integration step of 2 fs and holonomic constraints on all hydrogen-heavy atom bond terms were used. The energy of the system was minimized using conjugate-gradient minimization for 500 steps. Random velocities from a Maxwell distribution with T = 298.15 K were assigned to atoms. Then, the system was equilibrated first for 100 ps in the NVE ensemble and then for 1 ns in the NPT ensemble. In the latter case, temperature and pressure were controlled using the Langevin thermostat with a damping constant of 1 ps−1 and Berendsen barostat with a relaxation time of 400 fs, respectively. Finally, a 1 μs trajectory was calculated in the NVT ensemble using the Langevin thermostat with a damping constant of 0.1 ps−1. Trajectories were processed and analysed using the MDAnalysis Python package74.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
Protein structure data have been deposited in the PDB with accession codes: 7NYK (JIP1-SH3), 7NZB (JIP1-SH3(V517L)), 7NYO (JIP1-SH3(A541L)), 7NYL (JIP1-SH3(H493A)), 7NYM (JIP1-SH3(V517A)), 7NZC (POSH-SH3-1) and 7NZD (POSH-SH3-4). The 1H, 13C and 15N chemical shifts of JIP1-SH3 have been deposited in the Biological Magnetic Resonance Data Bank with accession codes: 50814 (JIP1-SH3), 50817 (JIP1-SH3(Y526A)), 50816 (JIP1-SH3(V517A)), 50818 (JIP1-SH3(H493A)) and 50815 (JIP1-SH3(A541L)). SH3 domain structures for molecular replacement were retrieved from the PDB (https://www.ebi.ac.uk/pdbe/) with accession codes: 2FPE (JIP1), 2LJ1 (SORBS1) and 2SRC (tyrosine protein kinase C-Src). Structures for the proteome-wide SH3 sequence analysis were retrieved from the PDB with accession codes: 1CSK, 3A98, 2O9S, 5VEI and 4LNP.
References
-
Campbell, I. D., Dobson, C. M. & Williams, R. J. Proton magnetic resonance studies of the tyrosine residues of hen lysozyme-assignment and detection of conformational mobility. Proc. R. Soc. B. 189, 503–509 (1975).
-
Wüthrich, K. & Wagner, G. NMR investigations of the dynamics of the aromatic amino acid residues in the basic pancreatic trypsin inhibitor. FEBS Lett. 50, 265–268 (1975).
-
Wagner, G., DeMarco, A. & Wüthrich, K. Dynamics of the aromatic amino acid residues in the globular conformation of the basic pancreatic trypsin inhibitor (BPTI). I. 1H NMR studies. Biophys. Struct. Mech. 2, 139–158 (1976).
-
Perutz, M. F. The role of aromatic rings as hydrogen-bond acceptors in molecular recognition. Philos. Trans. R. Soc. A 345, 105–112 (1993).
-
Brandl, M., Weiss, M. S., Jabs, A., Sühnel, J. & Hilgenfeld, R. C–H…π-interactions in proteins. J. Mol. Biol. 307, 357–377 (2001).
-
Steiner, T. & Koellner, G. Hydrogen bonds with π -acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 305, 535–557 (2001).
-
Burley, S. K. & Petsko, G. A. Aromatic–aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 (1985).
-
McGaughey, G. B., Gagné, M. & Rappé, A. K. π-Stacking interactions. Alive and well in proteins. J. Biol. Chem. 273, 15458–15463 (1998).
-
Dougherty, D. A. Cation–π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science 271, 163–168 (1996).
-
Ma, J. C. & Dougherty, D. A. The cation–π interaction. Chem. Rev. 97, 1303–1324 (1997).
-
Wagner, G. & Wüthrich, K. Dynamic model of globular protein conformations based on NMR studies in solution. Nature 275, 247–248 (1978).
-
Wagner, G. Activation volumes for the rotational motion of interior aromatic rings in globular proteins determined by high resolution 1H NMR at variable pressure. FEBS Lett. 112, 280–284 (1980).
-
Skalicky, J. J., Mills, J. L., Sharma, S. & Szyperski, T. Aromatic ring-flipping in supercooled water: implications for NMR-based structural biology of proteins. J. Am. Chem. Soc. 123, 388–397 (2001).
-
Hattori, M. et al. Infrequent cavity-forming fluctuations in HPr from Staphylococcus carnosus revealed by pressure- and temperature-dependent tyrosine ring flips. Protein Sci. 13, 3104–3114 (2004).
-
Rao, D. K. & Bhuyan, A. K. Complexity of aromatic ring-flip motions in proteins: Y97 ring dynamics in cytochrome c observed by cross-relaxation suppressed exchange NMR spectroscopy. J. Biomol. NMR 39, 187–196 (2007).
-
Weininger, U., Respondek, M., Löw, C. & Akke, M. Slow aromatic ring flips detected despite near-degenerate NMR frequencies of the exchanging nuclei. J. Phys. Chem. B 117, 9241–9247 (2013).
-
Weininger, U., Modig, K. & Akke, M. Ring flips revisited: 13C relaxation dispersion measurements of aromatic side chain dynamics and activation barriers in basic pancreatic trypsin inhibitor. Biochemistry 53, 4519–4525 (2014).
-
Yang, C.-J., Takeda, M., Terauchi, T., Jee, J. & Kainosho, M. Differential large-amplitude breathing motions in the interface of FKBP12–drug complexes. Biochemistry 54, 6983–6995 (2015).
-
Kasinath, V., Fu, Y., Sharp, K. A. & Wand, A. J. A sharp thermal transition of fast aromatic-ring dynamics in ubiquitin. Angew. Chem. Int. Ed. 54, 102–107 (2015).
-
Gauto, D. F. et al. Aromatic ring dynamics, thermal activation, and transient conformations of a 468 kDa enzyme by specific 1H-13C labeling and fast magic-angle spinning NMR. J. Am. Chem. Soc. 141, 11183–11195 (2019).
-
Dreydoppel, M., Raum, H. N. & Weininger, U. Slow ring flips in aromatic cluster of GB1 studied by aromatic 13C relaxation dispersion methods. J. Biomol. NMR 74, 183–191 2020).
-
Loria, J. P., Rance, M. & Palmer, A. G. A relaxation-compensated Carr–Purcell–Meiboom–Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 (1999).
-
Ishima, R. & Torchia, D. A. Extending the range of amide proton relaxation dispersion experiments in proteins using a constant-time relaxation-compensated CPMG approach. J. Biomol. NMR 25, 243–248 (2003).
-
Hansen, D. F., Vallurupalli, P. & Kay, L. E. An improved 15N relaxation dispersion experiment for the measurement of millisecond time-scale dynamics in proteins. J. Phys. Chem. B 112, 5898–5904 (2008).
-
Yuwen, T. & Kay, L. E. Revisiting 1HN CPMG relaxation dispersion experiments: a simple modification can eliminate large artifacts. J. Biomol. NMR 73, 641–650 (2019).
-
Sugase, K., Dyson, H. J. & Wright, P. E. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 (2007).
-
Baldwin, A. J. & Kay, L. E. NMR spectroscopy brings invisible protein states into focus. Nat. Chem. Biol. 5, 808–814 (2009).
-
Bouvignies, G. et al. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature 477, 111–114 (2011).
-
Neudecker, P. et al. Structure of an intermediate state in protein folding and aggregation. Science 336, 362–366 (2012).
-
Alderson, T. R. & Kay, L. E. Unveiling invisible protein states with NMR spectroscopy. Curr. Opin. Struct. Biol. 60, 39–49 (2020).
-
Kristensen, O. et al. A unique set of SH3–SH3 interactions controls IB1 homodimerization. EMBO J. 25, 785–797 (2006).
-
Dunbrack, R. L. & Karplus, M. Conformational analysis of the backbone-dependent rotamer preferences of protein sidechains. Nat. Struct. Biol. 1, 334–340 (1994).
-
Teyra, J. et al. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non-canonical specificities. Structure 25, 1598–1610 (2017).
-
Gehmlich, K. et al. Paxillin and ponsin interact in nascent costameres of muscle cells. J. Mol. Biol. 369, 665–682 (2007).
-
Hanawa-Suetsugu, K. et al. Structural basis for mutual relief of the Rac guanine nucleotide exchange factor DOCK2 and its partner ELMO1 from their autoinhibited forms. Proc. Natl Acad. Sci. USA 109, 3305–3310 (2012).
-
Borchert, T. V., Mathieu, M., Zeelen, J. P., Courtneidge, S. A. & Wierenga, R. K. The crystal structure of human CskSH3: structural diversity near the RT-Src and n-Src loop. FEBS Lett. 341, 79–85 (1994).
-
Chan, A. W. E., Hutchinson, E. G., Harris, D. & Thornton, J. M. Identification, classification, and analysis of β-bulges in proteins. Protein Sci. 2, 1574–1590 (1993).
-
Weininger, U., Respondek, M. & Akke, M. Conformational exchange of aromatic side chains characterized by L-optimized TROSY-selected 13C CPMG relaxation dispersion. J. Biomol. NMR 54, 9–14 (2012).
-
Weininger, U., Brath, U., Modig, K., Teilum, K. & Akke, M. Off-resonance rotating-frame relaxation dispersion experiment for 13C in aromatic side chains using L-optimized TROSY-selection. J. Biomol. NMR 59, 23–29 (2014).
-
Li, H., Yamada, H. & Akasaka, K. Effect of pressure on the tertiary structure and dynamics of folded basic pancreatic trypsin inhibitor. Biophys. J. 77, 2801–2812 (1999).
-
Dreydoppel, M., Dorn, B., Modig, K., Akke, M. & Weininger, U. Transition-state compressibility and activation volume of transient protein conformational fluctuations. JACS Au 1, 833–842 (2021).
-
Hart, D. J. & Tarendeau, F. Combinatorial library approaches for improving soluble protein expression in Escherichia coli. Acta Crystallogr. D 62, 19–26 (2006).
-
LeMaster, D. M. & Kushlan, D. M. Dynamical mapping of E. coli thioredoxin via 13C NMR relaxation analysis. J. Am. Chem. Soc. 118, 9255–9264 (1996).
-
Solyom, Z. et al. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J. Biomol. NMR 55, 311–321 (2013).
-
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
-
Jung, Y.-S. & Zweckstetter, M. Mars—robust automatic backbone assignment of proteins. J. Biomol. NMR 30, 11–23 (2004).
-
Marsh, J. A., Singh, V. K., Jia, Z. & Forman-Kay, J. D. Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation. Protein Sci. 15, 2795–2804 (2006).
-
Farrow, N. A. et al. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 (1994).
-
Vallurupalli, P., Bouvignies, G. & Kay, L. E. Studying ‘invisible’ excited protein states in slow exchange with a major state conformation. J. Am. Chem. Soc. 134, 8148–8161 (2012).
-
Christou, N. E. & Brutscher, B. BEST and SOFAST experiments for resonance assignment of histidine and tyrosine side chains in 13C/15N labeled proteins. J. Biomol. NMR 72, 115–124 (2018).
-
Yamazaki, T., Forman-Kay, J. D. & Kay, L. E. Two-dimensional NMR experiments for correlating 13Cβ and 1Hδ/ε chemical shifts of aromatic residues in 13C-labeled proteins via scalar couplings. J. Am. Chem. Soc. 115, 11054–11055 (1993).
-
Miloushev, V. Z. & Palmer, A. G. R1ρ relaxation for two-site chemical exchange: general approximations and some exact solutions. J. Magn. Reson. 177, 221–227 (2005).
-
Madeira, F. et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641 (2019).
-
Metsalu, T. & Vilo, J. ClustVis: a web tool for visualizing clustering of multivariate data using principal component analysis and heatmap. Nucleic Acids Res. 43, W566–W570 (2015).
-
Dar, I., Bonny, C., Pedersen, J. T., Gajhede, M. & Kristensen, O. Crystallization and preliminary crystallographic characterization of an SH3 domain from the IB1 scaffold protein. Acta Crystallogr. D 59, 2300–2302 (2003).
-
Gabadinho, J. et al. MxCuBE: a synchrotron beamline control environment customized for macromolecular crystallography experiments. J. Synchrotron Radiat. 17, 700–707 (2010).
-
Oscarsson, M. et al. MXCuBE2: the dawn of MXCuBE collaboration. J. Synchrotron Radiat. 26, 393–405 (2019).
-
Svensson, O., Malbet-Monaco, S., Popov, A., Nurizzo, D. & Bowler, M. W. Fully automatic characterization and data collection from crystals of biological macromolecules. Acta Crystallogr. D 71, 1757–1767 (2015).
-
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
-
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D 67, 293–302 (2011).
-
Monaco, S. et al. Automatic processing of macromolecular crystallography X-ray diffraction data at the ESRF. J. Appl. Crystallogr. 46, 804–810 (2013).
-
Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D 67, 282–292 (2011).
-
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204–1214 (2013).
-
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
-
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
-
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011).
-
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011).
-
Morin, A. et al. Collaboration gets the most out of software. eLife 2, e01456 (2013).
-
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
-
Wagner, J. R. et al. POVME 3.0: software for mapping binding pocket flexibility. J. Chem. Theory Comput. 13, 4584–4592 (2017).
-
Harvey, M. J., Giupponi, G. & Fabritiis, G. D. ACEMD: accelerating biomolecular dynamics in the microsecond time scale. J. Chem. Theory Comput. 5, 1632–1639 (2009).
-
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
-
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–28 (1996).
-
Michaud-Agrawal, N., Denning, E. J., Woolf, T. B. & Beckstein, O. MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 32, 2319–2327 (2011).
-
Dosset, P., Hus, J. C., Blackledge, M. & Marion, D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 (2000).
-
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Acknowledgements
We thank W. Adamski, S. Milles and T. Herrmann for discussions. We acknowledge the platforms of the Grenoble Instruct European Research Infrastructure Consortium (Integrated Structural Biology Grenoble; UAR 3518 CNRS-CEA-UGA-EMBL) within the Grenoble Partnership for Structural Biology. Platform access was supported by French Infrastructure for Integrated Structural Biology (ANR-10-INBS-05-02) and the Grenoble Alliance for Integrated Structural and Cell Biology, a project of the University Grenoble Alpes graduate school (Ecoles Universitaires de Recherche) CBH-EUR-GS (ANR-17-EURE-0003). The Institut de Biologie Structurale acknowledges integration into the Interdisciplinary Research Institute of Grenoble. The authors thank the Diamond Light Source and the European Synchrotron Radiation Facility for beamtime access and technical support. This work was funded by the Fondation pour la Recherche Medicale through a postdoctoral fellowship to L.M.P. (contract SPF201909009258) and by a PhD fellowship to J.K. from the IDPbyNMR Marie Curie action of the European commission (contract no. 264257). Funding is also acknowledged from the Agence National de la Recherche (ANR) through ANR T-ERC MAPKassembly (to M.R.J.), ANR ScaffoldDisorder (to M.R.J. and A.P.) and ANR JCJC RC18114CC NovoTargetParasite (to A.P.).
Author information
Affiliations
Contributions
L.M.P., M.R.J. and A.P. conceived the study. L.M.P., L.M.B., D.M. and J.K. made samples. L.M.B. and A.P. solved the structures of the POSH SH3 domains. L.M.P., F.S.I. and A.P. solved the structures of JIP1-SH3 and its variants. L.M.P., J.K. and M.R.J. designed and performed all NMR experiments. L.M.P., G.B. and M.R.J. analysed and interpreted the NMR data. M.B. and N.S. performed and analysed the MD simulation. M.R.J. and A.P. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Hashim Al-Hashimi and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Structural propensities and conformational exchange in JIP1-SH3.
a, 1H–15N HSQC spectrum of JIP1-SH3 at 25 °C with labels indicating assignments. Horizontal lines connect side chain resonances. b, Secondary structure propensities of JIP1-SH3 calculated from experimental 13Cα and 13Cβ chemical shifts at 25 °C. The position of secondary structure elements is indicated, as observed in the crystal structure of JIP1-SH3 (PDB: 2FPE). c, Differential scanning fluorimetry (DSF) melting curve for JIP1-SH3 (RFU – relative fluorescence unit). d, Conformational exchange contributions, REX, extracted from 15N CPMG relaxation dispersion data acquired at 15 °C of JIP1-SH3 (blue – 850 MHz, red – 600 MHz) and JIP1-SH3(Y526A) (orange – 700 MHz) as the difference between R2eff at low (31 Hz) and high (1 kHz) CPMG frequencies. e, Exchange contributions, REX, extracted from 1HN CPMG relaxation dispersion data acquired at 15 °C of JIP1-SH3 (blue – 950 MHz, red – 600 MHz) and JIP1-SH3(Y526A) (orange – 600 MHz) as the difference between R2eff at low (50 Hz) and high (2 kHz) CPMG frequencies. f, Structure of the dimeric SH3 domain of JIP1 with Y526 shown in cyan. Residues with exchange contributions (15N REX > 3 s−1 and 1HN REX > 10 s−1) are shown as spheres with colours indicating their position in the primary sequence (region 1 – green, region 2 – red, region 3 – blue). The residues showing 15N and 1HN exchange contributions are represented separately on each monomer.
Extended Data Fig. 2 Lipari–Szabo model-free analysis of 15N relaxation data of JIP1-SH3.
a, Region of the 1H–15N HSQC spectrum of JIP1-SH3 at temperatures ranging from 5 to 46 °C. b, Experimental 15N R2 (CPMG) relaxation rates at a 1H frequency of 600 MHz and four different temperatures. c, Experimental 15N R1 relaxation rates at a 1H frequency of 600 MHz and four different temperatures. d, Experimental {1H}-15N heteronuclear NOEs acquired at a 1H frequency of 600 MHz and 25 °C. Error bars in b–d represent one standard deviation (s.d.) derived from Monte Carlo simulations of experimental uncertainty. e, A model-free analysis of 15N R1, R2 and heteronuclear NOEs at 25 °C was carried out providing an axially symmetric diffusion tensor (Supplementary Discussion). The diffusion tensor is represented relative to the dimeric structure of JIP1-SH3. Distributions of axis orientations are shown as red dots and were determined from Monte Carlo simulations using Tensor275. f, Angular dependence of the R2/R1 ratios relative to the main axis of the diffusion tensor of JIP1-SH3. Only residues without exchange contributions to the transverse relaxation and for which the {1H}-15N NOE is above 0.7 were included in the analysis. Error bars are centred at experimental values and were propagated from the experimental uncertainty on R2 and R1. Orange squares are back-calculated values using the optimal tensor. g, Order parameters, S2, derived from the model-free analysis of the relaxation data at 25 °C. Error bars represent one standard deviation (s.d.) derived from Monte Carlo simulations as implemented in Tensor2. h, Conformational exchange contributions, REX, derived from the model-free analysis of the relaxation data at 25 °C. Green bars indicate residues that are located in the dimer interface of the SH3 domain as detected by the PISA server76.
Extended Data Fig. 3 Structure and dynamics of the Y526A variant of JIP1-SH3.
a, Comparison of the crystal structure of JIP1-SH3(Y526A) (green) with the WT structure (grey) showing an almost identical backbone conformation of the two proteins. The zoom highlights a minor structural difference at the level of the side chain of Q520 which reorients in the variant to take up the position normally occupied by Y526 in the WT protein. b, Superposition of 1H–15N HSQC spectra of JIP1-SH3(Y526A) (red) and JIP1-SH3(WT) (blue) acquired at 25 °C. c, Comparison of dihedral angles in JIP1-SH3(WT) (grey spheres) and JIP1-SH3(Y526A) (red lines). Dashed lines correspond to the backbone φ angle and full drawn lines to the backbone ψ angle. d, Conformational exchange contributions, REX, extracted from 15N CPMG relaxation dispersion data as the difference between R2eff at low (31 Hz) and high (1 kHz) CPMG frequencies. The exchange contributions are compared for WT JIP1-SH3 (grey, at 600 MHz) and the Y526A variant (red, at 700 MHz). e, Conformational exchange contributions, REX, extracted from 1HN CPMG relaxation dispersion data as the difference between R2eff at low (50 Hz) and high (2 kHz) CPMG frequencies. The exchange contributions are compared for the WT JIP1-SH3 (grey, at 600 MHz) and the Y526A variant (red, at 600 MHz). f–i, Chemical shift differences between the Y526A variant and WT JIP1-SH3 for 1HN (f), 15N (g), 13Cα (h) and 13Cβ (i).
Extended Data Fig. 4 Analysis of 15N and 1HN relaxation dispersion data of JIP1-SH3.
a, Examples of 15N CPMG relaxation dispersion profiles for JIP1-SH3 obtained at two magnetic field strengths (red – 600 MHz, blue – 850 MHz) at 15 °C. b, Examples of 1HN CPMG relaxation dispersion profiles for JIP1-SH3 obtained at two magnetic field strengths (red – 600 MHz, blue – 950 MHz) at 15 °C. The 15N and 1HN data were analysed simultaneously for all residues according to a two-site exchange model (full-drawn lines in a and b). Error bars in a and b represent one standard deviation (s.d.) derived from Monte Carlo simulations of experimental uncertainty. c, d, Chemical shift differences between the major and minor state extracted from a simultaneous analysis of the relaxation dispersion data at 15 °C for 15N (c) and 1HN (d).
Extended Data Fig. 5 Analysis of the sequence composition of 320 human SH3 domains.
a, Distribution of amino acid types at the position of Y526 of JIP1-SH3 following a sequence alignment of all 320 SH3 domains. b, Sequence alignment of all identified SH3 domains carrying either a phenylalanine (F) or tyrosine (Y) at the position of Y526 in JIP1-SH3. For each sequence the PCA group is indicated along with the PDB code and resolution of available crystal structures. c, Pie charts showing the distribution of amino acid types in the different SH3 domains at positions corresponding to residue 493, 517, 520 and 541 in JIP1-SH3. The pie charts are colour-coded according to the size score assigned to each amino acid type corresponding to the number of heavy atoms in their side chains (see online Methods). d, Illustration of the backbone conformation of the β-sheet formed between the 516–521 and 524–529 regions in SH3 domains with tyrosine or phenylalanine at position 526. The β-bulge conformation observed in WT JIP1-SH3 is observed in all SH3 domains for which high-resolution crystal structures are available.
Extended Data Fig. 6 Comparison of the structures of JIP1-SH3 and different variants.
a, Comparison of the crystal structures of JIP1-SH3(WT) (grey) and its V517L variant (green). The backbone conformation is entirely conserved with only minor rearrangements of protein side chains. b, Comparison of the crystal structures of the H493A (grey) and V517A (green) variants of JIP1-SH3. Structural features are conserved including similar side chain conformations. c, Comparison of the crystal structures of JIP1-SH3(WT) (grey) and its A541L variant (green) with arrows indicating the major conformational rearrangements between the WT protein and the variant. d, Illustration of the backbone conformation of the β-sheet formed between the 516–521 and 524–529 regions in the WT protein (left), and in the A541L variant (right). Dashed lines indicate hydrogen bonds. The A541L variant does not adopt a β-sheet conformation owing to the absence of several hydrogen bonds between the two β-strands. e, Schematic representation of the conformation of the β-strand encompassing residues 516 to 521 showing the orientation of the carbonyl group (“out” – carbonyl group surface exposed, “in” – carbonyl group pointing towards the β-strand encompassing residues 524 to 529) in the WT protein (left) and in the A541L variant (right). f, Volume of the Y526 pocket along the structural trajectory between the WT conformation and the conformations of the H493A/V517A mutants (grey) or A541L mutant (green). Conformations from the two structural trajectories at the maximum pocket volume are superimposed and shown in cartoon representation. The initial pocket expansion and its associated structural features are shared among all mutants.
Extended Data Fig. 7 Analysis of CPMG relaxation dispersion data of the H493A variant of JIP1-SH3.
a, Region of the 1H–15N HSQC of the H493A variant at three different temperatures (green - 15 °C, blue - 25 °C and red - 35 °C). b, Conformational exchange contributions, REX, extracted from 15N CPMG relaxation dispersion data of the H493A variant as the difference between R2eff at low (31 Hz) and high (1 kHz) CPMG frequencies (600 MHz and 15 °C). c, Conformational exchange contributions, REX, extracted from 1HN CPMG relaxation dispersion data of the H493A variant as the difference between R2eff at low (50 Hz) and high (2 kHz) CPMG frequencies (600 MHz and 15 °C). d, e, Examples of 15N (d) and 1HN (e) CPMG relaxation dispersion profiles of the H493A variant of JIP1-SH3 obtained at two magnetic field strengths (red – 600 MHz, blue – 950 MHz) at 15 °C. The 15N and 1HN data were analysed simultaneously for all residues according to a two-site exchange model (full-drawn lines in d and e) using a population of the minor state fixed to 10%. Error bars represent one standard deviation (s.d.) derived from Monte Carlo simulations of experimental uncertainty. f, g, Comparison of the chemical shift differences between the major and minor state extracted from relaxation dispersion experiments for WT JIP1-SH3 (red) and its H493A variant (blue). Data are shown for both 15N (f) and 1HN (g) chemical shifts.
Extended Data Fig. 8 Analysis of CPMG relaxation dispersion data of the V517A variant of JIP1-SH3.
a, Region of the 1H–15N HSQC of the V517A variant at three different temperatures (green - 15 °C, blue - 25 °C and red - 35 °C). b, Conformational exchange contributions, REX, extracted from 15N CPMG relaxation dispersion data of the V517A variant as the difference between R2eff at low (31 Hz) and high (1 kHz) CPMG frequencies (600 MHz and 15 °C). c, Conformational exchange contributions, REX, extracted from 1HN CPMG relaxation dispersion data of the V517A variant as the difference between R2eff at low (50 Hz) and high (2 kHz) CPMG frequencies (600 MHz and 15 °C). d, e, Examples of 15N (d) and 1HN (e) CPMG relaxation dispersion profiles of the V517A variant of JIP1-SH3 obtained at two magnetic field strengths (red – 600 MHz, blue – 950 MHz) at 15 °C. The 15N and 1HN data were analysed simultaneously for all residues according to a two-site exchange model (full-drawn lines in d and e) using a population of the minor state fixed to 11%. Error bars represent one standard deviation (s.d.) derived from Monte Carlo simulations of experimental uncertainty. f, g, Comparison of the chemical shift differences between the major and minor state extracted from relaxation dispersion experiments for WT JIP1-SH3 (red) and its V517A variant (blue). Data are shown for both 15N (f) and 1HN (g) chemical shifts.
Extended Data Fig. 9 The A541L variant does not capture the structural details of the minor state detected by CPMG relaxation dispersion in JIP1-SH3.
a, Superposition of 1H–15N HSQC spectra of JIP1-SH3(A541L) (red) and WT JIP1-SH3 (blue) acquired at 15 °C. b, Conformational exchange contributions, REX, extracted from 15N CPMG relaxation dispersion data of the A541L variant as the difference between R2eff at low (31 Hz) and high (1 kHz) CPMG frequencies at 700 MHz and 15 °C. Only modest 15N conformational exchange contributions are observed suggesting that this variant is populating a single conformation in solution represented by the determined crystal structure. c, d, Comparison of the chemical shift differences between the major and minor states extracted from relaxation dispersion experiments of WT JIP1-SH3 (blue) and the chemical shift differences between the observed chemical shifts of the A541L variant and WT JIP1-SH3 (red). Data are shown for 15N (c) and for 1HN (d) nuclei. The poor agreement between the two datasets show that the A541L crystal structure is not representative of the conformation of the minor state detected by NMR relaxation dispersion. e, f, Chemical shift differences between the A541L variant and WT JIP1-SH3 for 13Cα (e) and 13Cβ (f).
Extended Data Fig. 10 The aromatic ring of Y526 undergoes fast flipping.
a, 1H–13C HSQC spectra of JIP1-SH3 showing the region of tyrosine epsilon correlations. A sample of JIP1-SH3 site-selectively labelled at the epsilon positions was used, and spectra at four different temperatures were acquired in the range from 5 to 45 °C. b, Per-residue root-mean square fluctuation (RMSF) of Cα atoms during the MD simulation. c, Residue-specific Ramachandran plots showing the conformational sampling of β-strand residues during the MD simulation. The MD values are colour-coded from white to black according to simulation time. Blue regions indicate the allowed and marginally allowed regions. Red points indicate the starting conformation (one for each monomer of JIP1-SH3). Panels b and c demonstrate that the protein maintains a stable conformation throughout the MD simulation, while displaying several 180° ring flipping events of Y526 (Fig. 4d).
Supplementary information
Supplementary Information
This file contains the Supplementary Discussion (sections 1–3), Supplementary Fig. 1 and Supplementary Table 1.
Supplementary Video 1
Structural rearrangements associated with the transition between the major and minor state of JIP1-SH3. The animation shows how the major state of Y526 is stabilized by CH–π interactions from V517, Q520 and A541 and π–π interactions from H493. The transition to the minor state is associated with major structural rearrangements in the region encompassing residues 517 to 522. In particular, a classic β-bulge conformation is observed at residue 518 in the major state, which transitions to a canonical β-strand in the minor state. The video shows how these structural changes allow L519 to stabilize the ring of Y526 through CH–π interactions in the minor state.
Supplementary Video 2
Protein breathing motions generate a void volume around Y526 allowing fast ring flipping. The animation (surface representation) shows how a substantial void volume is generated around the tyrosine ring of Y526 through protein breathing motions along the structural trajectory from the major to the minor state. This cavity expansion allows fast time-scale ring flipping events of Y526 before closing the pocket of Y526.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mariño Pérez, L., Ielasi, F.S., Bessa, L.M. et al. Visualizing protein breathing motions associated with aromatic ring flipping. Nature (2022). https://ift.tt/87u0V9v
-
Received:
-
Accepted:
-
Published:
-
DOI: https://ift.tt/87u0V9v
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
"aromatic" - Google News
February 16, 2022 at 11:12PM
https://ift.tt/orywhck
Visualizing protein breathing motions associated with aromatic ring flipping - Nature.com
"aromatic" - Google News
https://ift.tt/PaRFkfI
https://ift.tt/YUHoGX7
Bagikan Berita Ini

















0 Response to "Visualizing protein breathing motions associated with aromatic ring flipping - Nature.com"
Post a Comment